ABRomics is an online community-driven platform to scale up and improve surveillance and research on antibiotic resistance from a One Health perspective.
Loading
ABRomics is an online community-driven platform to scale up and improve surveillance and research on antibiotic resistance from a One Health perspective.
UC4 : antimicrobial resistance analysis in the context of pulmonary infections.

Members
Coordinator :
G. Héry-Arnaud, UMR-1078/CBAM (Brest), Bacteriology; Project leader (BEACH study)
Team members :
O. Barraud, UMR-S1092 (Limoges), Bacteriology
L. Velo Suarez UMR-1078/CBAM (Brest)
A. Bihouée, BIRD platform (Nantes), Bioinformatics
S. Chaffron, LS2N-COMBI (Nantes), Bioinformatics
P. Plésiat, NRC Antibiotic Resistance – Pseudomonas (Besançon), Bacteriology
J. Poschmann, CRTI (Nantes), Bioinformatics
A. Roquilly, EA 3826 (Nantes), Intensive Care Medicine; Project leader (HAP2 and COVARDS studies)
Objective and outcome
This use case aims to demonstrate the contribution of the ABRomics platform in the analysis of antimicrobial resistance in the context of pulmonary infections
Outcome: ARGs expansion in the lung under antimicrobial selection pressure.
The use case is presented using the example of 2 types of lung infection corresponding to 2 different clinical and ecological contexts:
1° Chronic lung infections that require repeated and long-lasting antibiotic courses. The use of the ABRomics platform will be illustrated through the BEACH study (funding: PHRC-I API18/B/072 + Vaincre la Mucoviscidose), a prospective multicentre (n=11 centres in France) study (P.I. G. Héry-Arnaud) which aims to characterise the pulmonary and intestinal microbiome of newborns with cystic fibrosis (CF). The longitudinal metagenomic data generated by this program will enable the deciphering of the evolution of the bronchopulmonary community-acquired resistome during the first antibiotic cures (oral and inhaled routes) received by CF children in the first 2 years of life.
2° Acute lung infections, which are infections with a vital prognosis in the short term and which require rapidly effective antibiotic cures (short-lasting antibiotic courses, mostly intravenously). The use of the [name] platform will be illustrated through the HAP2 study (funding: H2020 #847782), a prospective international study on hospital-acquired pneumonia led by an international consortium (P.I. A. Roquilly), and the COVARDS study (funding ANR Flash-COVID), a French cohort of patients with severe COVID-19 pneumonia. Samples and raw data from these studies are already available. Multi-omic approach that integrates host-microbiota interactions generated by this program will enable the characterisation of the pulmonary ARGs distribution of hospital-acquired pneumonia (HAP) in intubated adult patients in a nosocomial context (HAP2) and in community-acquired pneumonia (COVARDS).
Pseudomonas aeruginosa is one of the most frequent and threatening bacteria linked with these both clinical entities, and belongs to the WHO global priority list of antibiotic-resistant bacterial species. Multidrug-resistant P. aeruginosa (MDR-PA) is an important and growing issue in the care of patients with CF. In VAPs, a high attributable mortality is observed despite adequate antimicrobial treatment that is increased in the presence of MDR-PA strains. The Pseudomonas NRC (headed by P. Plésiat) is currently developing a program to establish ARGs from clinical strains of different sources (including VAP and CF) and with well-defined antibiotypes. These WGS-derived data will enable PA-specific ARGs to be identified in the metagenomic data generated by the BEACH and HAP2 studies.
The three projects (BEACH, HAP2, COVARDS) will address specific lung AMR issues that will be resolved by the ABRomics PF.
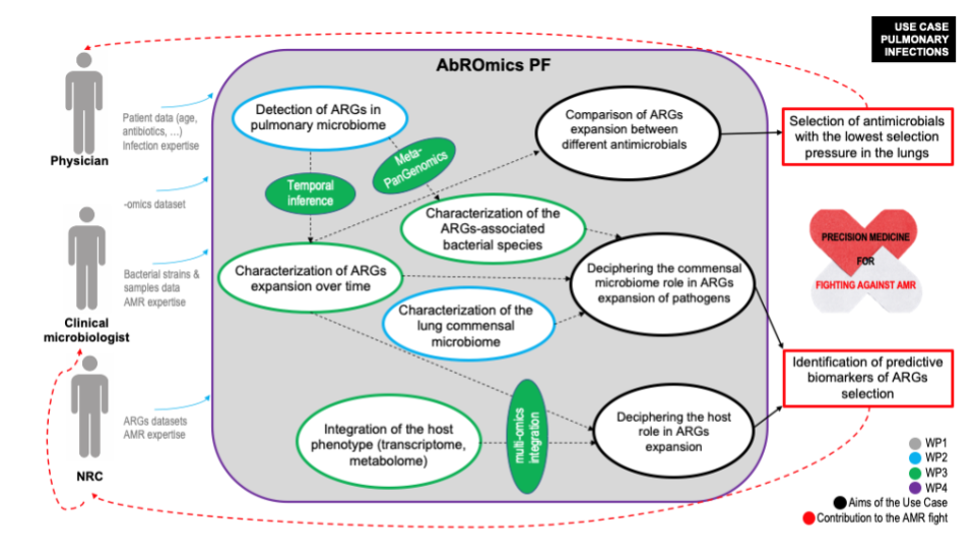
Lung infection use case diagram describing how physicians, microbiologists and National Reference Centres (NRC) will find the support of the ABRomics-PF to integrate their data and answer the 3 essential questions (circled in black) brought by the BEACH, COVARDS and HAP2 studies on the issue of antimicrobial resistance specific to the pulmonary ecosystem
Deliverable 1: lung microbiome characterization
Deliverable 2: lung host transcriptome and metabolome characterization
Deliverable 3: ARGs characterization of antimicrobial-naïve lungs in children and adult patients
Deliverable 4: ARGs characterization after lung antimicrobial courses
Deliverable 5: ARGs expansion in the lung under antimicrobial selection pressure
Deliverable 6: benchmarking of MustARD database and PCM workflow in the lung ecosystem
Deliverable 7: P. aeruginosa ARGs characterization from lung clinical strains isolated in different epidemiological contexts (community- or hospital-acquired infections)
Deliverable 8: lung commensals ARGs characterization
Deliverable 9: benchmarking of Meta-PanGenomics Graph application in the lung ecosystem
Deliverable 10: development of an ARGs database and workflows specific to the lung ecosystem taking into account the temporal and multi-omics scales