ABRomics is an online community-driven platform to scale up and improve surveillance and research on antibiotic resistance from a One Health perspective.
ABRomics is an online community-driven platform to scale up and improve surveillance and research on antibiotic resistance from a One Health perspective.
UC5 : One Health dissemination of antimicrobial resistance through aquatic environments

Members
Coordinators :
Anne-Laure Bañuls, MIVEGEC-IRD team (Montpellier) & Christian Lienhardt, TransVIHMI-IRD team (Montpellier) & Benoit Doublet, PGBA-INRAE team (Tours
Team members :
Mallorie Hide, MIVEGEC-IRD team (Montpellier)
Sylvain Godreuil, Bacteriology laboratory CHU Arnaud de Villeneuve/MIVEGEC-IRD team (Montpellier)
Juliette Hayer, MIVEGEC-IRD team (Montpellier)
Patrick Durand, MIVEGEC-IRD team (Montpellier)
Marion Vittecoq, Tour du Valat/MIVEGEC-IRD (Montpellier)
Patricia Licznar, HSM-IRD team (Montpellier)
Estelle Bilak, HSM-IRD team (Montpellier)
Fabien Aujoulat, HSM-IRD team (Montpellier)
François Sabot, MSO-IRD team (Montpellier)
Jean Christophe Auguet, MARBEC-IRD team (Montpellier)
Thierry Bouvier, MARBEC-IRD team (Montpellier)
Marine Combes, ISEM-IRD team (Montpellier)
Samira Serter, ISEM-IRD team (Montpellier)
Rudy Gonzlan, ISEM-IRD team (Montpellier)
Marie-Cécile Ploy, RESINFIT-INSERM team (Limoges)
Philippe Glaser, EERA-Institut Pasteur team (Paris)
Sébastien Leclercq, PGBA-INRAE team (Tours) Lulla Opatowski, EMEA-Institut Pasteur (Paris)
Objective and outcome
This use case aims to explore the role of different aquatic environments (waste water, continental and oceanic) in the dissemination of the antimicrobial resistance (AMR) thanks to the contribution of the ABRomics platform.
Outcome: Guidelines for targeted and/or shotgun multi-omic integration and application to ABR dissemination within and between aquatic ecosystems.
The use case is presented using the AMR data obtained from oceanic (MARBEC-IRD team), continental (irrigation scheme, underground water and rivers, freshwater aquaculture) (MIVEGEC, HSM, ISEM-IRD team; RESINFIT-INSERM team; PGBA-INRAE team) and wastewater (MIVEGEC, TransVIHMI-IRD team; RESINFIT-INSERM team; PGBA-INRAE team). Other public data sources may be used if relevant.
1) Oceanic waters
In the context of several projects (such as EXPLOI, PLASTIPATH and NYMPHE), samples were collected from various sources (water, sediments, fishes and plastics) to determine the circulation of AMR in oceanic environments. Plastics are included as they harbour pathogens known to be involved in animal and human epidemics. Thus, plastics and their associated antibiotic resistant pathogens in oceanic ecosystems can be transferred to marine animals ingesting plastics, and from them ultimately to humans who consume them.
Using the ABRomics platform, metagenomics and genomics data obtained from these samples will be confronted with various parameters (e.g. nutrient concentrations, temperature, salinity, antibiotic concentrations, precipitations), spatial (distance to the coast) and sociologic parameters (density of population, density of aquaculture exploitations) including local, regional and global human and animal AMR data to evaluate the determinants and risks of transmission in these environments.
2) Continental waters
The goal here is to investigate the contribution of continental waters to the transmission and dissemination of AMR and its impact on human, animal and environmental health (ZAARHYPOTTER project). In addition, we aim to understand the selection factors and spread of AMR in irrigation, drainage systems and aquaculture farms that can carry various kinds of pollutants (Antibiotic Resistance Genes – ARGs, antibiotic resistant bacteria – ARB, antibiotic residues and metals (MIVEGEC project, Bac Hung Gai project, TRANS-One Health project, WarmAqua). The sampling will be multiple: water, sediments, biofilms and sea fauna (fishes, shrimps, etc.). In these projects, we have collected microbiological data, metagenomics (bacterial diversity and resistome) and genomics (resistant bacterial genomes associated with resistance mechanisms) data as well as various parameters such as physical, geographical, climatic and chemical (natural and pollutant chemical composition) factors. The analysis of data according to the various parameters and the confrontation with local, regional and global AMR data using the ABRomics platform will help determine the risk factors for the detection and dissemination of AMR in these environments.
3) Waste waters
In the context of multicentre projects (CIRCUS, AMR Sud AVIESAN project (in review); ARCAHE, FSPI project, PRE-EMPT, TRANS-One Health project), we are – and will be – collecting wastewater samples together with human, animal and environmental (upstream/downstream rivers, soils) samplings. The overall objective is to characterize the circulation of AMR between human, animal and environmental compartments within the same living space, using a “One health” approach. The specific objectives are : (i) to characterize the resistance profiles of Enterobacteriaceae in these different compartments using culturomics, metagenomics and genomics; (ii) to determine and compare the proportion of multiresistant Enterobacteriaceae (MRE) in humans, animals and in waste waters; (iii) to characterize and compare the genome of MRE isolated in each compartment in order to identify the likely pathways of AMR circulation and identify the role of waste water in this transmission; and (iv) to describe the bacterial diversity and resistome through metagenomics. These data will be analysed together with data collected in humans and animals, and confronted to local, regional and global data available on AMR.
The project will address the role and contribution of water environments to the AMR, particularly in the dissemination of ARGs and ARB that will be available through the ABRomics PF:
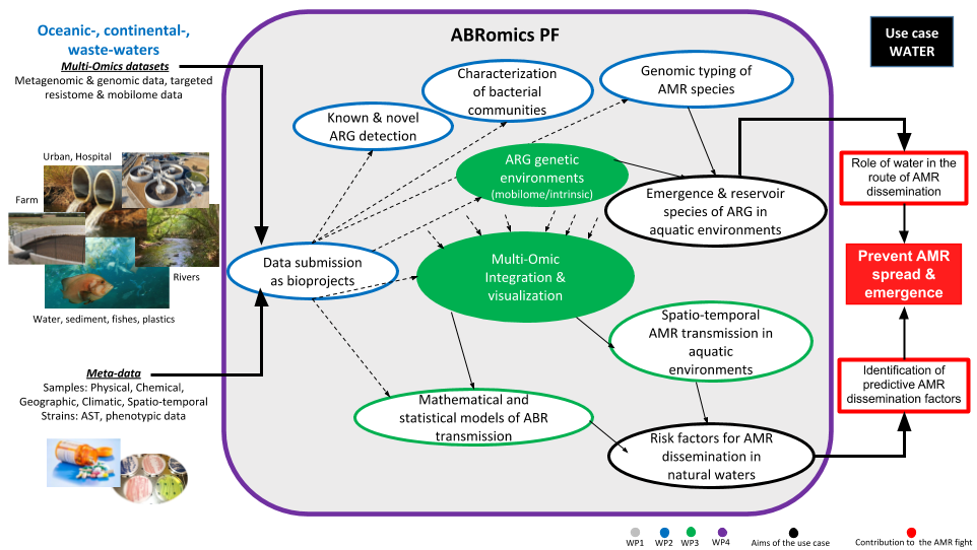
General diagram of bioproject data processing on the ABRomics-PF for several research studies related to AMR dissemination in aquatic environments. Data types, interactions with the ABRomics-PF components and scientific objectives to be reached and global outcomes are indicated.
Deliverable 1: water bacterial population characterization in the different water samples (water fauna, sediments, water, plastics) and ecosystems (oceanic, continental and waste waters);
Deliverable 2: water bacterial resistome characterization in the different water samples (water fauna, sediments, water, plastics) and ecosystems (oceanic, continental and waste waters)
Deliverable 3: bacterial genome and resistance mechanisms characterization
Deliverable 4: ARGs and enterobacterial genomes distribution according to the different water samples (water fauna, sediments, water, plastics) and ecosystems (oceanic, continental and waste waters)
Deliverable 5: potential role of water in the dissemination of AMR by comparison with global AMR data
Deliverable 6: benchmarking of MustARD database and PCM workflow in the lung ecosystem
Deliverable 7: development of an ARGs and MRE database and workflows specific to the water ecosystems considering the spatial scales and water characteristics