ABRomics is an online community-driven platform to scale up and improve surveillance and research on antibiotic resistance from a One Health perspective.
ABRomics is an online community-driven platform to scale up and improve surveillance and research on antibiotic resistance from a One Health perspective.
The informatics development of ABRomics platform are organized around four technical work packages:
– WP1. Computing architecture, aiming to provide the ABRomics consortium with the computing infrastructure and software environment.
– WP2. Integrated multi-omics microbiological database aiming to collect, standardize, organize, integrate and give access to the different types of multi-omics data relevant to ABR and perform standard automatic analysis on this data.
– WP3. Mathematical and bioinformatics tool development aiming to provide new mathematical and bioinformatics tools dedicated to (i) ABR multi-omics data integration, (ii) decipher mechanisms of ABR transmission (iii) model ABR emergence, selection and spread.
– WP6. Web portal open access, privacy-preserving Web tool to submit data, run programs and access results, transversal to all the other workpackages.
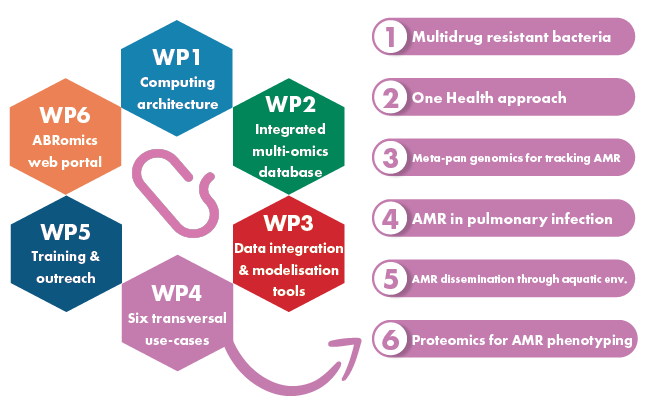
In order to ensure that the platform will respond to the needs of the communities, this development will be performed considering six transversal use cases (WP4).
The communication across the diversity of actors of ABR and towards the general public, together with training activities will be essential for the success of ABRomics (WP5).