ABRomics is an online community-driven platform to scale up and improve surveillance and research on antibiotic resistance from a One Health perspective.
ABRomics is an online community-driven platform to scale up and improve surveillance and research on antibiotic resistance from a One Health perspective.
UC1 : Detection and investigation of outbreak involving multidrug resistant bacteria

Members
Coordinator :
P. Plésiat, NRC Antibiotic Resistance (Besançon), Bacteriology
Team members :
A. Birer, NRC Antibiotic Resistance (Clermont-Ferrand), Bioinformatics
R. Bonnet, NRC Antibiotic Resistance (Clermont-Ferrand), Bacteriology
R. Beyrouthy, NRC Antibiotic Resistance (Clermont-Ferrand), Bacteriology
P. Lehours, NRC for Campylobacters and Helicobacters (Bordeaux), Bacteriology
Q. Jehanne, NRC for Campylobacters and Helicobacters (Bordeaux), Bioinformatics
P. Glaser, Unité Ecologie et Evolution de la Résistance aux Antibiotiques, Institut Pasteur (Paris), Bacteriology
Objective and outcome
The goal of this use case is to demonstrate the contribution of the ABRomics platform in the investigation of hospital or veterinary clinic outbreaks involving multidrug resistant (MDR) Gram-negative pathogens.
Outcome: guidelines and reproductible workflows to decipher the spatio-temporal dissemination of epidemic multidrug-resistant bacteria
Some mdr strains are able to actively diffuse within human populations, within animal populations, or both. Some can also persist in environmental reservoirs, from which patients or animals can be contaminated. Because of this complex landscape, identification of the transmission pathways that eventually lead to small or large infection outbreaks may be challenging, especially when involving clones able to survive or thrive in multiple ecosystems. Spatio-temporal analysis of the diffusion of such clones is therefore essential to implement appropriate control measures. Given that antibiotic resistance can be transmitted among bacteria through horizontal gene transfers, mobile genetic elements such as plasmids and ICEs (Integrative Conjugative Elements) must also be included in epidemiological surveys.
1. Transmissible clones of Escherichia coli, Klebsiella pneumoniae, Enterobacter cloacae, Acinetobacter baumannii, and Pseudomonas aeruginosa as well as Campylobacters and Helicobacter pylori are prone to accumulate resistance mechanisms up to becoming refractory to most antibiotic treatments. These so-called “high-risk” clones need to be detected and their diffusion carefully surveyed at the local (healthcare facilities), regional, national and international levels. Through cgMLST, and SNP analyses, the ABRomics platform will efficiently compare and dissect the genome sequences of clinical and environmental strains isolated during presumed hospital or veterinary clinic outbreaks, or exhibiting particular resistance traits (e.g., carbapenem or colistin resistance). The platform will be open to single users (e.g., medical biologists, researchers) or institutions (e.g., NRCs) with access to metagenomic data depending upon the authorization level. Based on these inputs, an interactive map of emergent or known “high-risk” clones will be drawn in real-time, and a warning made for confirmed cases of outbreak.
2. Self-transmissible and mobilizable plasmids carrying relevant ARGs (e.g., encoding carbapenemase, ESBL or colistin resistance) need to be traced in order to document their prevalence and diffusion across bacterial strains or species. The plasmid content of mdr strains of E. coli, K. pneumoniae, E. cloacae, A. baumannii, P. aeruginosa and Campylobacters will thus be systematically characterized (e.g., plasmid size, genes, incompatibility group, predicted transmissibility) from the genomic data submitted to the ABRomics platform. A comprehensive plasmid database will next be constructed in order to improve our knowledge of resistance replicons diffusing in different reservoirs and different geographic areas. Presence of specific virulence genes on plasmids will be screened and reported to the platform’s users.
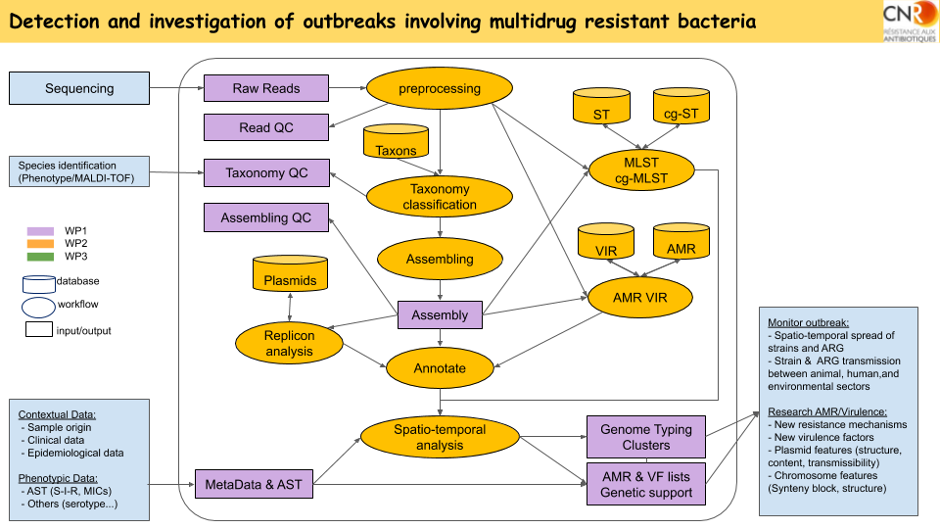
Workflow for sequencing data analysis in UC1 using ABRomics tools and DB
Deliverable 1: quality controls about the sequences, the assemblies
Deliverable 2: resistome and virulome of MDR strains
Deliverable 3: clonality relationships of MDR strains
Deliverable 4: spatio-temporal distribution of resistotypes, pathotypes and associated metadata
Deliverable 5: benchmarking about strain molecular typing and ABR gene detections