ABRomics is an online community-driven platform to scale up and improve surveillance and research on antibiotic resistance from a One Health perspective.
ABRomics is an online community-driven platform to scale up and improve surveillance and research on antibiotic resistance from a One Health perspective.

The ABRomics project aims to develop an online community-driven platform to scale up and improve surveillance and research on antibiotic resistance from a One Health perspective.
The integration and analysis of omics data with associated metadata requires the development of databases accessible to the community, the automatic use of standard pipelines for genomic and metagenomic data analysis, and the development of new bioinformatics and mathematical methods to answer generic and specific research questions related to AMR. There is also a need to develop a user-friendly Web interface for data deposit and for the presentation and retrieval of results.
The ABRomics platform is made of three essential components :
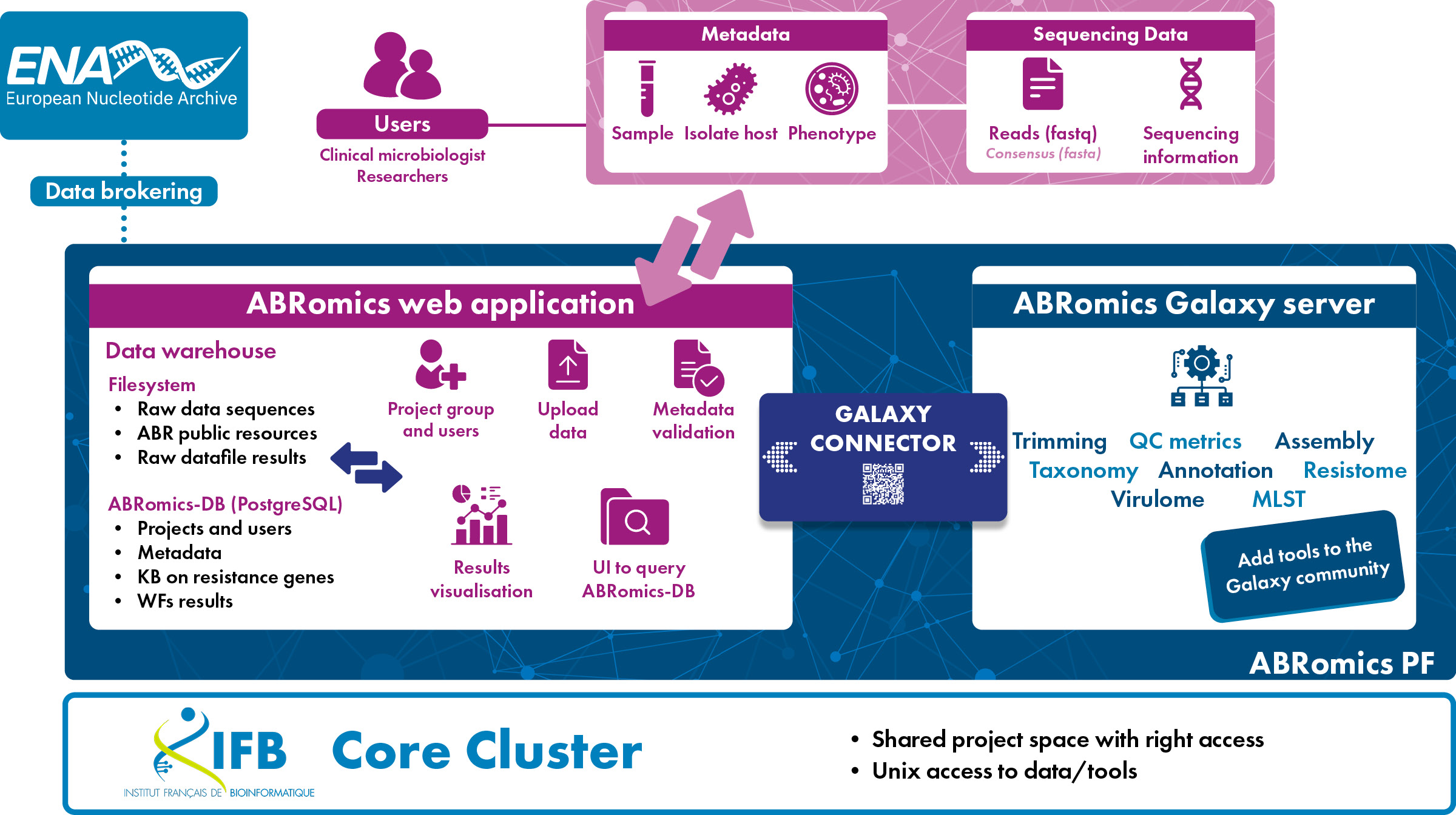
The three components of the ABRomics platform are running on the French Bioinformatics Institute (IFB) IT infrastructure: it offers high capacity of data storage and data analysis, accessible to users under defined rules (free to strict depending on the sensitivity of the data), and offering a general software environment, both HPC and Cloud environments.